CiteScore: 2.6, Q2 h-index: 30
CiteScore: 2.6, Q2 h-index: 30
Document Type : Cross Sectional Study
Authors
1 Department of Chemistry, Dr. D.Y. Patil Arts, Commerce & Science College; Pimpri, Pune-411018, India
2 Department of Chemistry, Dr. D.Y. Patil Arts, Commerce & Science Women College; Pimpri, Pune-411018, India
3 Department of Chemistry, Centre for Advanced Studies Department of Chemistry, Savitribai Phule Pune University, Pune (MH), India-411007
4 Department of Chemistry, Amruteshwar ACS, College, Vinzar, Pune (MH), India-412213
Graphical Abstract
Keywords
Subjects
Introduction
Indazole is an organic heterocyclic compound called isoindazole or benzo[c]pyrazole, 1,2-benzodiazole [1]. The discovery of indazolering was invented by Emil Fisher calling them fused benzene and pyrazole ring [2-3]. The continuous research on indazole is due to its interesting chemical and biological properties. Indazole belongs to azole family and the presence of 10π electron in the ring of indazole, makes them aromatic character similar to other heterocyclic aromatic compounds like indole, pyrimidine and pyrrole. The indazole exists in 1H-, 2H- and 3H- three tautomeric in nature (Figure 1) [4].
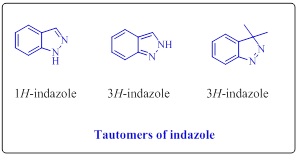
Figure 1: three tautomeric of indazole
In the case of the tautomeric equilibrium1H- and 2H-indazole, 1H tautomer is more stable than 2H tautomer. Tautomeric equilibrium stability was calculated by thermal and photochemical methods, for example, 2- methyl-2H-indazole is less stable than 1-methyl-1H-indazole. However 3H-indazole have only a few examples are known, which possess aliphatic or aromatic groups on the cyclic ring [5]. The example Pkb values for 1- methyl-1H-indazole is 0.42 but Pkb values for 2-methyl-2H-indazole is 2.02 that means the 2H tautomer is a stronger base than 1H tautomer. For identification of 1H tautomer and 2H tautomer the spectral studies 1H, 13C, 14N, and 15N NMR are used [6,7]. The 1HNMR signal for methyl protons 1-methyl and 2-methyl-indazole vary only by 0.1-0.2 ppm, while in 13C NMR the 1H tautomer is appear at 132-133 ppm and 2H tautomer has appeared at 123-124 ppm [8]. World’s largest trading drugs are nitrogen-containing heterocycles. Indazole heterocyclic has two nitrogen atoms in the five-member ring. It is because of their availability all over nature, they make them a key scaffold of many biological and medicinally important molecules [9-10]. Indazole moiety has some pharmaceutical, agrestic, and commercial applications [11]. Indazole moiety is a medicinally significant scaffold having the most prominent skeleton of wild range drug ligand, such as serotonin 5-HT₃ receptor antagonis [12] applied in the cancer therapy and benzyl amine a COX-inhibitor inhibitor [13] anti-tumour [14-15], anti-HIV [16], Antimicrobial [14], and strongly estrogen receptor [18]. This has aroused great interest in the development of novel indazole-based therapeutic agents. The isolation of indazole natural products is very rare and less [19]. During the last two decade there is VI natural products of indazole are isolated in the scientific community. The natural product such as Nigellicine [20] (1), Nigeglanine [21] (2), Nigellidine [22], Nigellidine-O-sulfite, [23] nigeglapine (5) and nigeglaquine [24] (6) (Figure 2).
The Nigellicine natural product of indazole was isolated during the era of 1994-1985 from the flowering plant Nigella sativa native. Thousands of years ago, the Nigella Sativa seeds were traditionally used for the cure of various infections [20].
In the pursuit of developing novel antimicrobial agents, herein we demonstrate the preparation of some novel hybrids indazole moiety to investigate their potential antimicrobial activities.
Our research group is always interested in finding new heterocyclic compounds for biological testing [25-30]. The main objective of our continuous research is to find out new leads molecules with potential antimicrobial activities. Considering the medicinal utility of indazole, we attempted the molecular docking of novel multi substituted indazole derivatives with attractive targets enzyme with Clorobiocin ligand [31].
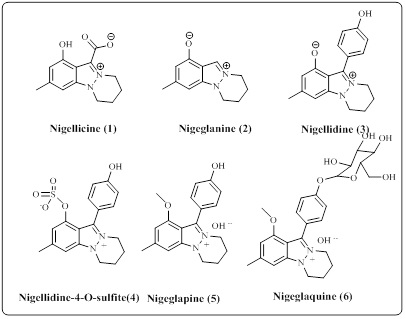
Figure 2: Natural products of Indazoles
Material and Methods
General Information
All the melting points were uncorrected and determined in an open capillary tube. The chemicals and solvents used in this study were of laboratory grade and purified. Completion of the reaction was monitored by thin layer chromatography on precoated sheets of silica gel-G (Merck, Germany) using iodine vapors for detection. IR spectra were recorded in KBr pellets on an FTIR Perkin Elmer/Schimadzu/Bruker spectrophotometer. 1H NMR (400 MHz) and 13C NMR (100 MHz) spectra were recorded in (DMSO)-d6with an Avance spectrometer (Bruker, Germany) at a 400-MHz frequency using TMS as an internal standard; chemical shifts are reported in parts per million and coupling constant in hertz (Hz). Multiplicities are reported as follows: s (singlet), d (doublet), t (triplet), m (multiplet). Mass spectra were recorded on an EI-Shimadzu QP 2010 PLUS GC-MS system (Shimadzu, Japan).
Experimental Section
General procedure for the Synthesis of Synthesis of 1,1'-(4-hydroxy-2-(1H-indol-3-yl)-4-methyl-6-oxocyclohexane -1,3-diyl) bis (ethan-1-one(4A);
The mixture of starting compound diketo compound and appropriate aldehyde in dimethylsulfoxide with base piperidine in a 50 mL round bottom flask. The reaction mixture was magnetically stirred at ambient temperature. The reaction mixture stirring for 5 h and completion of the reaction was checked by using TLC, after completion of the reaction, the reaction mixture was acidified with 10% HCl, quenched into ice-cold water, washed with water to afford the pure solid product
Light Yellow solid, M.P.97-98 °C, Yield : 98 % ; FTIR; 1690-1709, 2975, 2978, 2917, OH -3485, 1H NMR (300 MHz, CDCl3) δ 16.19 (s,1H), 8.25, (s, 1H), 7.84 (d, J = 2.8 Hz, 1H), 7.67 (d, J = 7.7 Hz, 1H), 7.39 – 7.32 (m, 2H), 7.22 –7.15 (m, 2H), 4.30 (t, J = 12.1Hz, 1H), 4.11 (s, 1H), 4.03 (d, J = 12.4 Hz, 1H), 3.60 (d, J = 11.8 Hz, 1H), 2.71- 2.57 (m, 2H), 2.00 (s, 3H), 1.62 (s, 3H), 1.33 (s, 3H),13CNMR; CHCl3, 217.32, 210.31, 109.72, 141.21, 130.15, 128.46, 126.13, 123.76, 123.59, 116.61, 77.71, 72.7, 67.26, 59.14, 36.60, 35.09, 33.08.Chemical Formula: C19H21NNaO4 Exact Mass: 350.1368, Obtained: 350.1360.
Synthesis of 1,1'-(2-(5-bromo-1H-indol-3-yl)-4-hydroxy-4-methyl-6-oxocyclohexane-1,3-diyl) bis (ethan-1-one) (4B)
Light white solid M.P.177-178 °C, (C19H20BrNO4), FT-IR; 1690-1714, 2985, 2970, 2917, OH -3490; 1HNMR (300 CDCl3); δ 4.204 (t, 1H, 4.302 s, 1H), 3.907 (d, 1H, J= 12.4), 3.702 (d,1H, J = 11.8), 2.591 (dd, 2H, J = 12.7 and 12.6 Hz), 2.01 (s, 3H), 1.623, s, 3H), 1.34 (s, 3H), δ 13CMR; (75 MHz CHCl3), 214.30, 210.35, 108.61, 138.23, 129.21,127.47, 124.16, 123.21, 121.52, 1151.6, 74.58, 73.62, 67.33, 59.42, 34.77, 33.23, 18.48.
Synthesis of 1,1'-(4-hydroxy-2-(5-methoxy-1H-indol-3-yl)-4-methyl-6-oxocyclohexane-1,3-diyl) bis (ethan-1-one) (4C)
White solid, m.p. 145-147 °C, M.F. C20H23NO5) 1H NMR (300 MHz, DMSO) δ 10.78 (s, 1H), 7.46 – 7.03 (m, 3H), 6.73 (s, 1H 5.21 (s, 1H)), 4.26 (t, J = 11.6Hz, 1H), 4.10 (d, J = 11.9 Hz, 1Hz, 1H), 3.78 (s, 3H), 3.00 (d, J = 12.1 Hz, 1H), 2.35 (d, J =13.8 Hz, 1H), 1.89 (s, 6H), 1.18 (s, 3H).13CMR (75 MHz CHCl3); δ 210.50, 206.97, 204.87, 153.46, 122.72, 127.91, 27.82, 112.56, 111.2, 109.9, 104.5, 72.75, 56.00, 55.60,30.430.46, 28.58, 21.58, 15.21.
General procedure for the synthesis of 1-(4-(1H-indol-3-yl)-3,6-dimethyl-4,5-dihydro-1H-indazol-5-yl) ethan-1-one (5A)
A solution of compound 1,1'-(4-hydroxy-2-(1H-indol-3-yl)-4-methyl-6-oxocyclohexane -1,3-diyl) bis (ethan-1-one and hydrazine hydrate 2.1 Equiv. in methanol, reflux the reaction mixture, TLC show the completion of stating compound, cool the reaction and poured in ic cold water, filter the residue and dry under vacuum gives light yellow solid with 65 % yield.
Lemon solid, m.p. 165 °C (decomposed); FTIR, 3346, 3292,3047, 2968, 2922, 1693, 1641; 1H NMR (300 MHz, DMSO), δ 12.32 (s, 1H), 7.87 (d, J = 7.8 Hz, 1H), 7.42 (d, J = 8.3 Hz,1H), 7.20 (t, J = 7.4 Hz, 1H), 7.11 (t, J = 7.6 Hz, 1H), 5.33 (s, 1H), 4.20 (d, J = 12.2 Hz, 1H), 3.11 (d, J = 14.0 Hz, 1H), 2.44 (d, J = 14.0 Hz, 1H), 1.99 (s, 3H), 1.97 (s, 3H), 1.27 (s, 3H).
1-(6-hydroxy-4-(4-methoxyphenyl)-3,6-dimethyl-4,5,6,7-tetrahydro-1H-indazol-5-yl) ethan-1-one (5F)
Faint yellow solid, M.P. 182-185 °C, FTIR; 3412, 3057, 3001, 29682837, 1614, 1695, 1585, 1H NMR (300MHz, DMSO), δ 6.98 (d, J =Hz, 2H), 6.73 (d, J = 8.7Hz, 2H), 4.00 (d, J = 10.9 Hz, 1H), 3.69(s, 3H), 2.86 (d, J= 11.0 Hz, 1H), 2.71 (dd, J = 36.1, 16.5 Hz, 2H), 2.51 (s, 1H), 1.65 (s, 3H), 1.48 (s, 3H), 1.23 (s, 3H). δ 13C (75MHz, dmso) 216.56, 158.38, 133.86, 129.14, 114.04, 113.48, 71.63, 64.10, 55.11, 40.92, 36.66, 34.39, 28.63. M. F.C18H22N2O3, Exact Mass: 314.1630, obtained; 314.9.
1-(6-hydroxy-3,6-dimethyl-4-phenyl-4,5,6,7-tetrahydro-1H-indazol-5-yl) ethan-1-one (5D)
White solid, m.p. 184-186 °C; FT-IR; 3273, 3161, 3088, 2974,2914,1676, 1583 1H NMR (300MHz, DMSO) δ11.97 (s, 1H), 7.23 (dt, J = 18.2, 7.1 Hz, 5H), 4.63 (s, 1H), 4.19 (d, J = 10.9 Hz,1H), 2.89 – 2.39 (m, 4H), 2.00 (s, 3H), 1.40 (s, 3H), 1.21 (s, 3H) 13CNMR (75 MHz, DMSO), 209.56, 146.67, 141.56, 137.43, 128.54, 128.55, 128.18, 128.21, 126.10, 116.90, 74.34, 70.89, 41.22, 32.33, 31.62,24.55, 14.17, C17H20N2O2, mass found 284.1525, obtained m/z; 284.
1-(4-(4-chlorophenyl)-6-hydroxy-3,6-dimethyl-4,5,6,7-tetrahydro-1H-indazol-5-yl) ethan-1-one (5E)
Light yellow solid, m.p. 154-155°C, FTIR, 3404, 3311, 3059, 2928,1704, 1597, 1H NMR (300 MHz, CDCl3) δ7.28 (d, J = 8.3 Hz, 2H), 7.11 (d, J = 8.2 Hz, 2H), 4.15 (d, J = 10.9 Hz, 1H), 2.95(d, J = 11.0 Hz, 1H), 2.82 (dd, J = 41.9, 16.6 Hz, 2H), 1.75 (s, 3H), 1.59 (s, 3H), 1.35 (s, 3H). M.F. C17H19ClN2O2, Exact Mass: 318.1135, obtained LCMS, m+h; 319.0
1-(3,6-dimethyl-4-(1-methyl-1H-indol-3-yl)-4,5-dihydro-1H-indazol-5-yl) ethan-1-one (5H)
Cream color, m.p. 171-174 °C, FT-IR; 3691, 3398, 3119, 2968,2787, 1697, 1602, 1512, 1H NMR (300 MHz, DMSO) δ 8.66(s, 1H7.50 (d, J = 7.3 Hz, 2H), 7.35 (s, 1H), 7.22 (d, J = 5.1 Hz,2H),7.2– 7.13 (m, 1H), 7.04– 6.98 (m, 1H), 4.87 (s, 1H), 4.70 (d, J = 11.1 Hz, 1H), 3.82 (s, 3H), 3.11 (d, J = 11.3 Hz, 1H), 2.15 (s, 3H), 1.63 (s, 3H), 1.33 (s, 3H).
Results and Discussion
Initially, we started our strategy for the preparation of diacetyl, hydroxy-indole, methylcyclohexanone derivatives (Scheme 1), (4A-4C). In the present investigation, the synthesis of 2,4-diacetyl-5-hydroxy-3-(1H-indol-3-yl)-5-methylcyclohexanone analogs (Scheme 2, Figure 3) was performed by using modified aldol condensation reactions of heteroaromatic aldehydes and acetylacetone in DMSO solvent yielded intermediate cyclohexanones in excellent yields [32-33] (Scheme 2). When indole-3-carbaldehyde was stirred with acetylacetone in the presence of base heterocyclic amine (20 mol%) produce a precursor 1-3 analogs in excellent yields.
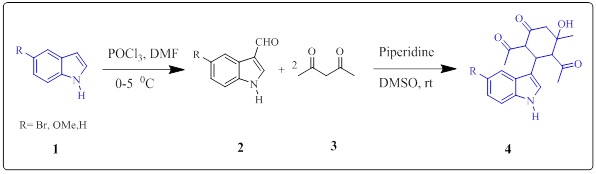
Scheme 1: Synthesis of multi - substituted cyclohexanone derivatives
All new compounds were characterized using 1H NMR, 13CNMR, IR, and MS spectroscopic techniques. In the IR spectrum of 1, the stretching band due to hydroxyl group was observed at 3485 cm-1 and the stretching frequency for carbonyl carbon C=O stretching was observed at 1690 and 1709 cm-1. The 1H NMR spectrum showed singlets for D2O exchangeable OH at 4.102for 1H proton and three singlets for three methyl groups each appear at 1.999 s, 3H, 1.617, s, 3H, 1.333, s. 3H. Moreover in cyclohexanone moiety, there are four types of proton appearing at δ4.304 triplet for 1H proton and 4.027, d, 1H, 3.602, d, 1H proton. The coupling constant of protons. H-2 and H- 4 is J = 12.6 and 11.7 Hz. Indicate trans to each other 13CMR chemical shifts of the C=O group show three downfield signals corresponding to two acetyl carbonyl groups and one cyclohexanone carbonyl group appear at 217.3, 210.3, 109.7 ppm. The compound formation was finally confirmed by HRMS, (M+Na) for Chemical Formula: C19H21NNaO4 calculated mass is 350.1368, obtained HRMS 350.1360.
Next, our strategic move towards the synthesis of 1-(4, 5, 6, 7-tetrahydro-6-hydroxy-4-(1H-indol-3-yl)-3,6-dimethyl-1H-indazol-5-yl). ethanone derivatives (Scheme 2, Figure 3). The cyclohexanone derivatives containing 1,3-dicarbonyl moiety reacted with hydrazine hydrate or phenylhydrazine hydrochloride in the acid medium resulted in the formation of 1-(4,5,6,7-tetrahydro-6-hydroxy-4-(1H-indol-3-yl)-3,6-dimethyl-1H-indazol-5-yl). ethanone derivatives derivative.
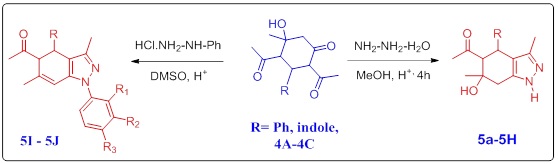
Scheme 2: Synthesis of tetrahydro-1H-indol-3-yl)-R-1H-indazol-5-yl) ethanone
Synthesized scaffolds were characterized by numerous spectroscopic techniques. In the IR spectrum of analogstetrahydro-1H-indazol-5-yl) ethan-1-one5A showed the IR signal appear at 3346 and 3292indicate presence of N-H and OH functional group. The IR signal that appears at 1693 is a stretching signal for the carbonyl group. In 1H NMR spectrum of product, 5A showed a signal at δ 12.32 (s, 1H) and 5.33 (s, 1H), corresponding to N-H, D2O exchangeable proton, and one singlet at δ5.55 s, 1H for OH, which is D2O exchangeable proton. One singlet at 6.73 s, 1H belongs to the proton of indole CH. There are three singlets for three CH3groupsthatappear at δ1.27, s, 3H, δ1.99 s, 3H, δ1.97 s, 3H. In 13C NMR the signal at 202.23 corresponds to an acetyl carbonyl group. Finally, in mass spectroscopy (ESI) m/z (M+H) calculated 323.1634observed at Exact Mass: 324.16 The product formation was established by 1H, 13C NMR analytical techniques. On the basis of characterization reports and their evaluation using numerous techniques, the structure of product 5A was assigned. The compound 5C showed the FTIR band at 3412, 3057 indicating the presence of Oh and N-H functionalities in the compound. The 1HNMR 1H NMR (300MHz, dmso) show the δ 6.98 (d, J = 8.7 Hz, 2H), 6.73 (d, J = 8.7Hz, 2H), 4.00 (d, J = 10.9 Hz, 1H), 3.69 (s, 3H), 2.86 (d, J = 11.0 Hz, 1H), 2.71 (dd, J = 36.1, 16.5 Hz, 2H), 2.51 (s, 1H), 1.65 (s, 3H), 1.48 (s, 3H), 1.23 (s, 3H). The 13C (75 MHz, DMSO) 216.56, 158.38, 133.86, 129.14, 114.04, 113.48, 71.63, 64.10, 55.11, 40.92, 36.66, 34.39, 28.63. M. F. C18H22N2O3, Exact Mass: 314.1630, obtained; 314.9. On the basis of the evaluation report using spectral techniques, the structure of scaffold 5C was assigned.
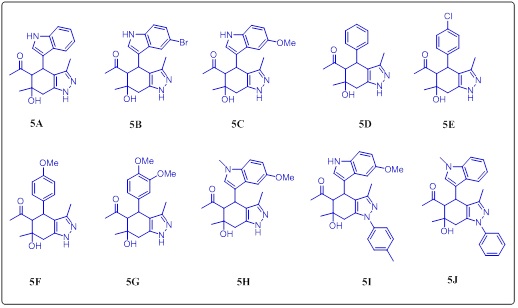
Figure 3: The physicochemical properties data ofmultisubstitutedindazole
Biology
Antibacterial Activity
The antibacterial activity of the synthesized compounds was performed using the Agar-diffusion protocol [35]. The obtained result is projected with the using zone of inhibition (ZOI) of the compound as well as their minimum inhibitory concentrations (MICs) values against the standard and blank sample. The concentration of the solution is diluted by dissolving 1-2 mg/mL series of the compound in a DMSO solvent and poured into the bacteria seeded agar plates. The activity of the compound was calculated by counting the zone of inhibition of the compound in mm and minimum inhibitory concentrations MICs (μg/mL).
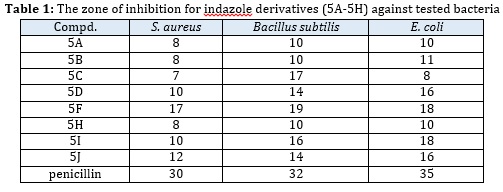
The obtained result was correlated with standard penicillin antibacterial drug (Table 1), herein we found that the scaffold 5A, 4D, and 5F display highest antibacterial activity (Table 1). While, in the case of scaffolds 5B, 5C, and 5H display good potential activity against the selective microorganism Escherichia Coli (MTCC- 443), Bacillus subtilis (MTCC-441), and Staphylococcus aureus (MTCC-96) (Table 1). All the other remaining molecules 5G, 5I, and 5Jrevealedminor antibacterial activity. It is concluded that as the concentration of compounds increases from 1 mg/mL to 2 mg/mL, the zone of inhibition also increases, hence at the higher concentration at 2.5 mg/mL the zone of inhibition is higher.
Molecular docking
From the literature data, it was found that the given compounds are active against the cancer research, therefore we decided to perform the molecular docking of 21-(6-hydroxy-4-(1H-indol-3-yl)-3,6-dimethyl-4,5,6,7-tetrahydro-1H-indazol-5-yl) ethan-1-one 5A-5J compound with 1kzn protein. In order to understand the nature of interactions of ligands, we have carried out molecular docking analysis by using Autodock software (Dallakyan S. et al. 2015). The 1kzn protein is known as DNA gyrase enzyme. The DNA gyrase is a most of the protein is the bacterial enzyme which takes part in the biochemical process like transcription and copying new stand, that enhances the formation of negative DNA supercoil of bacteria. The DNA gyrase is the representative target for the antibacterial agents it blocks the formation of new copying DNA, which results in bacterial death. The three-dimensional crystal structure of crystal structure of 1kzn was retrieved from the Protein Data Bank (http:// www.rscb.org/pdb). [31] The native ligand (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1Yl)Phenyl) Acrylic Acid (Azd9496) was deleted before CADD study. The active sites/ binding sites of the protein were identified using the Computed Atlas of Surface Topography of Proteins (Castro) program (http: //cast.engr.uic.edu) [34] The compound 5A-5J is well accommodated in the binding sites of 1KZN protein. While the compound 5A is also well accommodated in the binding sites of 1kzn protein and shows -7.9 kcal/mol binding energy and exhibit hydrogen bonding with Gly-77 amino acids and the molecule shows van der Waals of interaction with amino acid residues like FLU-50, ASP-4, ALA-47 ASN-46, THR-165, PRO-79, IIL- amino acids (Figure 4).
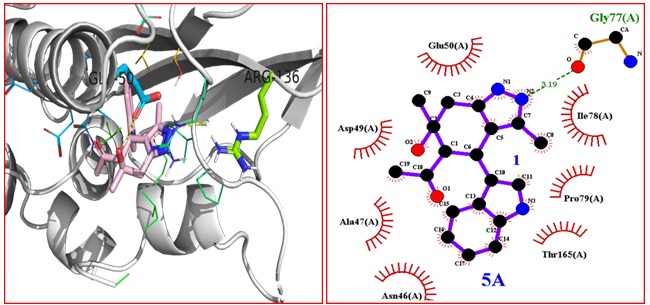
Figure 4: Binding interactions between 5A and DNA gyrase 10 (PDB code 1KZN). B) 2D structure showing hydrogen bonding and hydrophobic interactions of 5A with DNA gyrase (PDB code 1KZN) complex
The compound 5D is well accommodated in the binding sites of 1KZN protein. While the compound 5D is also well accommodated in the binding sites of 1kzn protein and shows -7.0 kcal/mol binding energy and exhibit hydrogen bonding with ARG-76, 2.98 GLU-50 2.85, GLY-77, 2.92 amino acids.
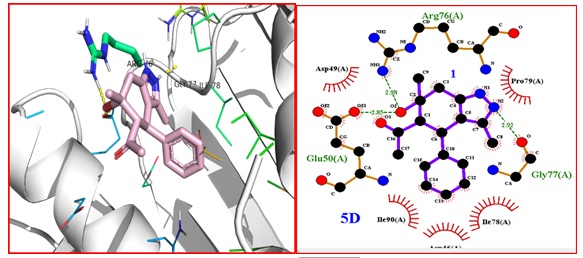
Figure 5: A). Binding interactions between 5D and DNA gyrase 10 (PDB code 1KZN). B) 2D structure showing hydrogen bonding and hydrophobic interactions of 5D with 9 DNA gyrase (PDB code 1KZN) complex
The ligand shows H-bond with ARG-76, having bond distance NH---O 2.98 Å, the amino acid GLU-50 show H- bond with H---OH having h-bond 2.85 Å and amino acid GLY-77, display H-bond with N---OH having distance 2.85 Å and the molecule shows van der Waals of interaction with amino acid residues like IlE-90, IlE-78, PRO-9, ASP-49 amino acids (Figure 5).
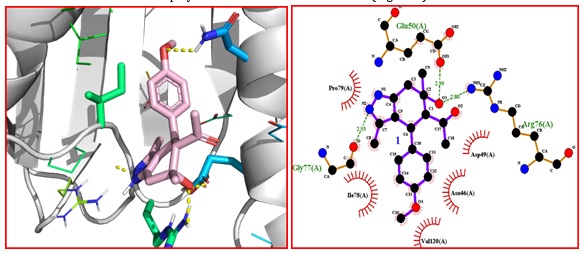
Figure 6: A). Binding interactions between 5F and DNA gyrase 10 (PDB code 1KZN). B) 2D structure showing hydrogen bonding and hydrophobic interactions of 5F with 9 DNA gyrase (PDB code 1KZN) complex
While the compound 5Fisalso well accommodated in the binding sites of 1kzn protein and shows -7.0 kcal/mol binding energy and exhibit hydrogen bonding with AGLU-50, 2.99a, GLY- 2.93A, ARG-76-2.80 amino acids. The ligand shows H-bond with GLU-50, having bond distance NH---O 2.99 Å, the amino acid GLY- 2.93A show H- bond with H---OH having h-bond 2.93 Å and amino acid ARG-76, display H-bond with N---OH having distance 2.80Å and the molecule shows van der Waals of interaction with amino acid residues like PRO-9, ILE-78, VAL-120, ASN-46, ASP-49, amino acids (Figure 6).
Conclusions
This study demonstrates a novel, efficient, method for the synthesis of novel series of 1-(3,6-dimethyl-4-(1-methyl-1H-indol-3-yl)-4,5-dihydro-1H-indazol-5-yl) ethan-1-one derivatives. The compound 5A, 4D, and 5F shows highest antibacterial activity and the remaining compounds exhibited moderate antibacterial activity against the S. aureus, Bacillus subtilis and E. Coli. Finally, the molecular docking studies releveled that the compound 5D and 5F posses excellent bonding interaction with the targeted enzyme with DNA gyrase (PDB code 1KZN).
Acknowledgments
M.V.G is thankful to Department of Chemistry, Dr. D.Y. Patil Arts, Commerce & Science College; Pimpri, Pune-411018 for research support. S.V.G is thankful to Department of Chemistry, Dr. D.Y. Patil Arts, Commerce & Science Women's College; Pimpri, Pune-411018 for research support
Authors' contributions
All authors have contributed significantly and met criteria for authorship. All the authors read and approved the final copy of the manuscript.
Conflict of Interest
We have no conflicts of interest to disclose.
HOW TO CITE THIS ARTICLE
Milind Gaikwad, Sunil Gaikwad, Rahul Kamble. Synthesis of Novel Series of 1-(6-hydroxy-4-(1H-indol-3-yl)-3,6-dimethyl- 4,5,6,7-tetrahydro-1H-indazol-5-yl) ethan-1-oneas evaluations of their antimicrobial activity with Insilco docking study, J. Med. Chem. Sci., 2022, 5(2) 239-248
DOI: 10.26655/JMCHEMSCI.2022.2.11
| Article View | 10,491 |
| PDF Download | 2,286 |