Document Type : Review Article
Authors
- Hoseinali Danesh 1, 2
- Saeid Rahimi Ghasabeh 3
- Ramin Rounasi 4
- Ehsan Zarepur 5
- Ali Abdolrazaghnejad 2, 6
1 Plastic, Reconstructive & Aesthetic Surgeon, Assistant Professor of Zahedan University of Medical Sciences (ZAUMS)
2 Clinical Immunology Research Center at Zahedan University of Medical Sciences, Zahedan, Iran
3 Anesthesiologist at Noor Afshar hospital, Iranian Red Crescent Society, Iran
4 Department of Anesthesiology and Pain Medicine, Iran University of Medical Sciences, Tehran, Iran
5 Resident of cardiology, Isfahan cardiovascular research center, Isfahan cardiovascular research institute, Isfahan University of Medical Sciences, Isfahan, Iran
6 Department of Emergency Medicine, Khatam-Al-Anbia Hospital, Zahedan University of Medical Sciences, Zahedan, Iran
Abstract
The aim of current study was to evaluate the outcome of some laboratory tests and accuracy of diagnostic tests for patients with COVID-19. The QUADAS-2 tool was utilized to quality assessment of diagnostic accuracy studies. For data extraction, two reviewers blind and independently extracted data from abstract and full text of studies that included. 95% confidence interval for effect size with fixed effect model and Inver-variance method were calculated. Meta-analysis was performed using Stata/MP v.16 software. 469 studies were reviewed, of which the full text of 52 studies was reviewed and finally twenty-two studies were selected for meta-analysis. Mean differences of Alanine transaminase was 3.40 U/L (MD, 95% CI -2.45, 9.25), correlation between Alanine transaminase and severe COVID-19 was not significantly. Stool, feces, rectal swabs 25% (ES, 95% CI 0.32, 0.82), Urine 1% (ES, 95% CI 0.05, 0.58), overall sensitivity of PCR for detection of COVID-19 was 44% (ES, 95% CI 0.19, 0.68), less sensitive observerd when PCR detection of COVID-19. Evidence revealed that at the time of admission of patients with COVID-19, a specific laboratory model can be used to perform relevant tests and make decisions about patients. PCR using sputum samples was highly sensitive for detecting COVID-19 and after that computed tomography of the chest was identified with high sensitivity.
Graphical Abstract
Keywords
Main Subjects
Introduction
Patients have reported early symptoms of fever and cough, and most have complained of chest pain, difficulty breathing, and pneumonia, which are diagnosed clinically by imaging tests such as chest x-rays or computed tomography (CT) scan [1-4]. Common findings of chest CT images in people with COVID-19 (Figure 1) are multifocal bilateral patchy ground-glass opacities or consolidation with interlobular septal and vascular in the peripheral areas of the lungs [5-8]. However, CT findings can change as the disease progresses, and the manifestations may be consistent with other viral respiratory illnesses [9-11]. Reverse transcription polymerase chain reaction (RT-PCR) is used to detect COVID-19 based on molecular testing, which aims to detect virus RNA in patient respiratory samples [12-14]. This test is time consuming and laboratory equipment shortages are common around the world, and if the viral genome is not enough, the result will be a false negative [15-17]. Other methods used to diagnose the disease include serological testing of IgM and IgG production in response to viral infection [18-20]. Most of these methods are used when the rate of asymptomatic infections increases or PCR testing is not available [21-23]. In the current study, tried to investigate the characteristics (sensitivity, specificity) of all COVID19 diagnostic tests [24-26]. On the other hand, laboratory data are very important in achieving important results. Therefore, the aim of current study was evaluated the outcome of Laboratory Tests and accuracy of diagnostic tests in Patients with COVID-19 [27-29].

Figure 1: Universities Create New Epidemiological Model to Study COVID-19 Dynamics
Material and Methods
Selection Criteria
Inclusion criteria: patients with COVID-19 infections; Sample size higher than 4 patients; f any diagnostic method; Human biological samples (HBS). Case studies, case reports and reviews; other infection were excluded from the study. Studies data were reported by study, years, study design, age, Number of patients, Methods for Evaluation. The QUADAS-2 tool was used to quality assessment of diagnostic accuracy studies [22]. It consists of four main domains comprising patient selection, index test, reference test, and flow/timing. All four domains include two independent judgments: “Risk of bias” and “Applicability”. Only the last domain “flow/timing” comprises a single judgment on “Risk of Bias”. All seven judgments must be scored “Yes” or “No” or “Unclear”. The purpose of the signalling questions is to help score the QUADAS criterion “Risk of bias” as “Low”, “High” or “Unclear”. For Data extraction, two reviewers blind and independently extracted data from abstract and full text of studies that included. Prior to the screening, kappa statistics was carried out in order to verify the agreement level between the reviewers. The kappa values were higher than 0.80. 95% confidence interval for mean difference with fixed effect model and Inverse-variance method were calculated. To deal with potential heterogeneity, random effects were used and I2 showed heterogeneity. I2 values less than 50% indicate low heterogeneity and above 50% indicate moderate to high heterogeneity. Meta-analysis was performed using Stata/MP v.16 software (The fastest version of Stata).
Results and Discussion
In the review of the existing literature using the studied keywords, 469 studies were found. In the initial review, duplicate studies were eliminated and abstracts of 434 studies were reviewed. At this stage, 361 studies did not meet the inclusion criteria, so they were excluded, and in the second stage, the full text of 73 studies was reviewed by two authors. At this stage, 52 studies were excluded from the study due to incomplete data, inconsistency of results in a study, poor studies, lack of access to full text, inconsistent data with the purpose of the study. Finally, twenty-two studies were selected (Figure 2).

Figure 2: Study Attrition
Characteristics
Twelve and ten studies have been included in present article for laboratory tests and outcome and accuracy of diagnostic tests, respectively.
The number of patients were 2314 with 43.75 mean of years. The range of radiological abnormalities at baseline was between 44-100%. The mean of comorbidity 36.11% (Table1).
The number of patients were 1662. Reference method in all studies was RT-PCR, one studies nor reported this section. In two studies methods for evaluation, enzyme linked immunosorbent assay (ELISA), in four studies Chest CT and in three studies RT-PCR were used. The marker gene is presented in Table 1.
Bias Assessment
Qu et al., 2020 [30] and Xie et al., 2020 [31] studies had a total score of 2 and 3/14, this score indicates the low quality of the studies. (0-4, high risk of bias). Eight studies had a total score between 5-9/14 (moderate risk of bias), this score indicates the moderate quality of the studies. Twelve studies had a total score between 10-14/14 (low risk of bias), this score indicates the high quality of the studies [32-35].

Outcome of Laboratory Tests
Mean differences of White blood cells in patients with COVID-19 was 1.8 ×109/L (MD, 95% CI 0.18, 1.98). Decrease the number of lymphocytes in the blood in patients with COVID-19 was -0.31×109/L (MD, 95% CI -0.41, -0.21). Increased number of neutrophils observed in patients with severe COVID-19, the Mean differences of this parameter was 1.31 ×109/L (MD, 95% CI 0.51, 2.11). A low hemoglobin count observed in patients with severe COVID-19, the Mean differences of this parameter was -4 g/L (MD, 95% CI -6.5, -1.5). The Mean differences of procalcitonin was 0.04 ng/ml (MD, 95% CI 0.00, 0.07), there was no association between procalcitonin and the severity of COVID-19. The range for prothrombin time was 0.31-1.11 seconds and Mean differences was 0.71 seconds, this means was prothrombin time was high in patients with COVID-19. Mean differences of creatinine in patients with COVID-19 was 3.20 µm (MD, 95% CI -0.70, 7.10), correlation between creatinine and severe COVID-19 was not significantly (Figure 3 and Table 4) [36-38].

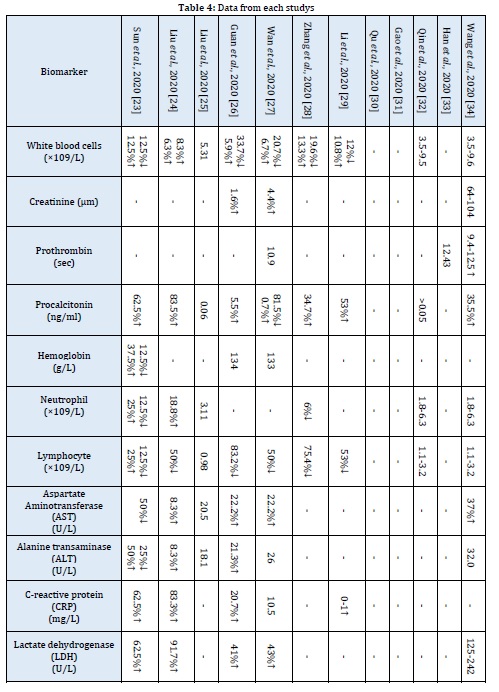
Outcome of Laboratory Tests
Mean differences of C-reactive protein was 49.10 mg/L (MD, 95% CI 31.10, 67.10), high level of CRP in the blood observed in patients with severe COVID-19. High Lactate dehydrogenase was observed in patients with severe COVID-19, the Mean differences of this parameter was 196 U/L (MD, 95% CI 130.50, 261.5). The Mean differences of Aspartate Aminotransferase was 8.30 U/L (MD, 95% CI 2.85, 13.75), this result showed small difference of Aspartate Aminotransferase in patients with severe COVID-19. Mean differences of Alanine transaminase was 3.40 U/L (MD, 95% CI -2.45, 9.25), correlation between Alanine transaminase and severe COVID-19 was not significantly (Figure 3 and Table 4) [39].

Accuracy of Diagnostic Tests
Meta analysis of Computed tomography sensitive was 92% (ES, 95% CI 48%, 100%) among five studies and heterogeneity found (I2=0%; P =1.00) (Low heterogeneity) (Figure 4). Meta analysis of Computed tomography specificity was 73% (ES, 95% CI 0.29, 1.16%) among five studies and heterogeneity found (I2=0%; P =0.87) (Low heterogeneity) (Figure 5 and 6).

Sub group Meta Analysis of Immunological Tests (IgM and IgG):
sensitivity of type of sample:
blood 82% (ES, 95% CI 0.13, 1.51), serum 82% ES, 95% CI 0.16, 1.80), overall sensitivity of Immunological tests was 82% (ES, 95% CI 0.25, 1.39), and heterogeneity found (I2=0%; P =1.00) (Low heterogeneity). Acording to Test of group differences there was no significant differences were observed between type of sample for detection of COVID-19 (p=1.00) (Figure6).
specificity of type of sample:
blood 91% (ES, 95% CI 0.21, 1.60), overall specificity of Immunological tests was 91% (ES, 95% CI 0.21, 1.60), and heterogeneity found (I2=0%; P =0.99) (Low heterogeneity) (Figure 7).

Figure 7: Forest plot showed sensitivity of Immunological tests (IgM and IgG) diagnostic for COVID-19

Figure 8: Forest plot showed specificity of Immunological tests (IgM and IgG) diagnostic for COVID-19
Sub group Meta analysis of PCR showed
Sensitivity of type of sample
Stool, feces, rectal swabs 25% (ES, 95% CI 0.32, 0.82), Urine 1% (ES, 95% CI 0.05, 0.58), Blood 7% (ES, 95% CI 0.06, 0.77), Nasopharyngeal aspirate, nasopharyngeal and throat swab 73% (ES, 95% CI 0.24, 1.24), Sputum 97% (ES, 95% CI 0.28, 1.66), Saliva 62% (ES, 95% CI 0.07, 1.31). overall sensitivity of PCR for detection of COVID-19 was 44% (ES, 95% CI 0.19, 0.68), less sensitive observerd when PCR detection of COVID-19 (Figure 9).
The findings of the present study were strong in relation to the first objective of the study (evaluated the outcome of Laboratory Tests) and no high heterogeneity was found between the studies. Present meta-analysis showed higher WBC count, neutrophils, prothrombin time; lower lymphocytes and hemoglobin count observed in patients with COVID-19 [4]. There was no correlation between procalcitonin; creatinine and severe COVID-19. The quality of the studies was moderate to high [5]. The results are consistent with common findings in other viral infections, with a recent systematic review showing that procalcitonin in the management of pneumonia cannot determine whether the infection is bacterial or viral [1]. The exact cause of death from COVID-19 is not yet known, but multiple organ dysfunction and hypoxia can be causes of death [5]. No correlation has been reported between renal function and disease severity at the time of admission [3]. In the present study, high quality studies were used and one of the strengths of their results was that laboratory analysis of patients with COVID-19 was reported at the time of admission. One of the limitations of the study was the lack of definition of the severity of the disease, the period of hospitalization, and follow-up. It is suggested that studies with different populations and comparison of laboratory results be performed in this field [2]. Also, in the present study, a study was conducted in children, which needs to be further studied in different age populations.
The findings of the present study were strong in relation to the second objective of the study (accuracy of diagnostic tests). High sensitivity and low specificity of computed tomography was obseverd; High Sensitivity and high specificity of Immunological tests was obseverd; low Sensitivity of PCR detection of COVID-19 was obseverd. The most important findings of the present study were that meta-analysis showed that among all the methods discussed, PCR using sputum samples was highly sensitive to the others methods and is one of the most important methods for detecting COVID-19. These results are supported by the CDC Recommendations, which state that initial testing for SARS-CoV-2 should prioritize the collection of respiratory samples [6]. Computed tomography the second test with high sensitivity was identified, although the specificity for this test was less. Demonstration in computed tomography of the chest is a very important strategy for complementary diagnosis. On the other hand, due to the lower sensitivity of serological tests than other methods, using serological tests to diagnose in the early / acute stage of the disease can be challenging. One study showed that remodeling of IgM and IgG can occur simultaneously or sequentially in COVID-19, and antibody titers reach 19 after 6 days [5]. It should be noted, however, that the strength of immunological testing is its rapid return time and relatively low cost, which may be an alternative due to the lack of RT-PCR and its high cost. One of the limitations of this study in examining the second goal is the weakness in the methodology of the studies and the lack of examination of the characteristics of patients and samples.

Figure 9: Forest plot showed sensitivity of PCR diagnostic for COVID-19
Conclusion
Evidence showed that at the time of admission of patients with COVID-19, a specific laboratory model can be used to perform relevant tests and make decisions about patients. The laboratory findings in patients with COVID-19 were almost identical to the results of routine laboratory tests for other viral infections. PCR using sputum samples was highly sensitive for detecting COVID-19 and after that computed tomography of the chest was identified with high sensitivity.
Acknowledgments
The authors express their gratitude to the Vice Chancellor for Research and Information Technology of Zahedan University of Medical Sciences for approving this research and the head of Khatam al-Anbia Hospital as well as all participating families for their cooperation in doing this research.
Future research
It is necessary to conduct further studies with larger sample sizes to investigate administering the right sedative for traumatic children who are admitted to the emergency department. It is also rewarding to use ketamine and midazolam in non-emergency situations where there is enough time to sedate the patient or in emergency departments that are not very crowded.
Funding
The current study was funded by Zahedan University of Medical Sciences.
Authors' contributions
All authors have contributed significantly and met criteria for authorship. All the authors read and approved the final copy of the manuscript.
Conflict of Interest
We have no conflicts of interest to disclose.
ORCID
Hoseinali Danesh:
https://www.orcid.org/0000-0002-0385-2597
Ali Abdolrazaghnejad:
https://www.orcid.org/0000-0002-4121-1643
HOW TO CITE THIS ARTICLE
Maryam Ziaei, Mahjoubeh Keykha, Faeze Kazemi, Ali Abdolrazaghnejad. Comparing the Sedative Effect of Oral Midazolam Versus Oral Ketamine on Children Aged 1-7 Years in Need of Radiologic Procedures, J. Med. Chem. Sci., 2022, 5(2) 227-238
DOI: 10.26655/JMCHEMSCI.2022.2.10
- Mokhtare M., Alimoradzadeh R., Agah S., Mirmiranpour H., Khodabandehloo N., Middle East J. Dig. Dis., 2017, 9:228 [Google Scholar], [Publisher]
- Etemadi S., Mahmoodiyeh B., Rajabi S., Kamali A., Milanifard M., Romanian Soc. Cell Biol., 2021, 25:2417 [Google Scholar], [Publisher]
- Amini A., Shahpoori Arani H., Milani Fard M., Eurasian J. Sci. Tech., 2021, 1:421 [Crossref], [Google Scholar], [Publisher]
- Givi F., Esmaeili R., Mojab F., Nasiri M., Shadnoush M., Koomesh, 2019, 21:254 [Google Scholar], [Publisher]
- Estebsari F., Dastoorpoor M., Khalifehkandi Z.R., Esmaeili R., Aghababaeian H., Aging Sci., 2020, 13:4 [Crossref], [Google Scholar], [Publisher]
- Mohammadi M., Esmaeili R., Fani M., Adv. Pharm. Educ. Res., 2019, 9:111 [Google Scholar], [Publisher]
- Sardari M., Esmaeili R., Ravesh N.N., Nasiri M., Adv. Pharm. Educ. Res., 2019, 9:145 [Google Scholar], [Publisher]
- Hajalimohammadi M., Esmaeili R., Zandi M., Zadeh B.P., Medico-Legal Update, 2020, 20:262 [Crossref], [Google Scholar], [Publisher]
- Azadmehr F., Esmaeili R., Farahani Z.B., Arabborzu Z., Adv. Pharm. Educ. Res., 2018, 8:1 [Google Scholar], [Publisher]
- Esmaeili R., Barziabadi Z.F., Khoob M.K., Nephro-Urol. Mon., 2021, 13:e100728 [Crossref], [Google Scholar], [Publisher]
- Soleimani F., Anbohi S.Z., Esmaeili R., Pourhoseingholi M.A., Borhani F., Clin. Diagn. Res., 12:LC01 [Crossref], [Google Scholar], [Publisher]
- Maddah Z., Ghalenoee M., Mohtashami J., Esmaieli R., Naseri-Salahsh V., J. Islam. Repub. Iran, 2018, 32:1 [Crossref], [Google Scholar], [Publisher]
- Bahoush G., Pahlavani R., Salarinejad S., Zarei E., PCR, 2021, 16: 166 [Crossref], [Google Scholar], [Publisher]
- Silva E.H., Lekamwasam S., Prev. Epidemiol., 2020, 5:e03 [Crossref], [Google Scholar], [Publisher]
- Alimoradzadeh R., Mokhtare M., Agah S., J. Age., 2017, 12:78 [Google Scholar], [Publisher]
- Alimoradzadeh R., Mirmiranpour H., Hashemi P., Pezeshki S., Salehi S.S., Neurology Neurophys., 2019, 10:1 [Google Scholar], [Publisher]
- Abdolrazaghnejad A., Banaie M., Safdari M., Ad. J. Emerg. med, 2018, 2:1 [Google Scholar], [Publisher]
- Akhlaghi N., Payandemehr P., Yaseri M., Akhlaghi A.A., Abdolrazaghnejad A., Emerg. Med., 2019, 73:462 [Crossref], [Google Scholar], [Publisher]
- Abdolrazaghnejad A., Banaie M., J.Pharma, 2017, 12:180 [Crossref], [Google Scholar], [Publisher]
- Pakniyat A., Qaribi M., Hezaveh DR., Abdolrazaghnejad A., Acute Dis, 2018, 7:241 [Crossref], [Google Scholar], [Publisher]
- Rahmati J., Fathi H., Sultanova N., Davudov M.M., Danesh HA., J. Otorhinolaryngol. Head Neck. Surg., 2020, 9:86 [Crossref], [Google Scholar], [Publisher]
- Rakei S., Rad H.I., Arbabisarjou A., Danesh H.A., Drug Invent. Today, 2019, 11: 3123 [Google Scholar], [Publisher]
- Rakei S., Rad H.I., Irandegani F., Danesh H.A., Drug Invent. Today, 2019, 12: 2809 [Google Scholar], [Publisher]
- Danesh H.A., Focus Med. Sci. J., 2018, 4 [Crossref], [Google Scholar], [Publisher]
- Danesh H.A., Saboury M., Sabzi A., Saboury M., Jafary M., Saboury S., J. Islam. Repub. Iran, 2015, 29:172 [Crossref], [Google Scholar], [Publisher]
- Abdolrazaghnejad A., Banaie M., Safdari M., J. Emerg. Med., 2018, 2:1 [Google Scholar], [Publisher]
- Akhlaghi N., Payandemehr P., Yaseri M., Akhlaghi AA., Abdolrazaghnejad A., Emerg. Medicine, 2019, 73:462 [Crossref], [Google Scholar], [Publisher]
- Abdolrazaghnejad A., Banaie M., J. Pharma, 2017, 12:180 [Crossref], [Google Scholar], [Publisher]
- Pakniyat A., Qaribi M., Hezaveh DR., Abdolrazaghnejad A., Acute Dis., 2018, 7:241 [Crossref], [Google Scholar], [Publisher]
- Mehr S.S., Ramezani A., Kashi M.A., Krimpalis S., J. Mat. Sci., 2018, 53:14629 [Crossref], [Google Scholar], [Publisher]
- Mehr SS., Ramezani A., Kashi M.A., J. Mat. Sci, Mat. Elect., 2018, 29:18771 [Crossref], [Google Scholar], [Publisher]
- Nickavar A., Abolhasan Choobdar F., Mazouri A., Talebi A., J. Neonatol. 2018, 9:1 [Crossref], [Google Scholar], [Publisher]
- Nejad N.H., Saboute M., Hosseini R., Tahoori M., Otukesh H., Comp. Pediat., 2019, 10:e74359 [Crossref], [Google Scholar], [Publisher]
- Hoseiny-Nejad , Cheraghi T., Nikpour S., Sheikhvatan M. I. Pedia., 2018, 28:e63588 [Crossref], [Google Scholar], [Publisher]
- Saboute , Mazouri A., Khalesi N., Hoseiny Nejad N., Razaghian A., Iran. J. Neonatol. 2017, 8:83 [Crossref], [Google Scholar], [Publisher]
- Hoseiny Nejad N., Sadat Sharif A., Otukesh H., Hekmat S., Sakhaei M., Pediatric Nephrol., 2021, 36:1 [Crossref], [Google Scholar], [Publisher]
- Bahoush G., Zarei E., Open Maced. J. Med. Sci., 2020, 8:233 [Crossref], [Google Scholar], [Publisher]
- Bidari A., Hassanzadeh M., Naderkhani M., Mesgarha M.G., Mohammad A.P., Azadeh A., Hossein H., Zarei E., Khodadost M., J. Islam. Repub. Iran., 2021, 35:1 [Crossref], [Google Scholar], [Publisher]
- Nazari Sabet M, Ahmadipour E, Zamansaraei S., Prev. Epidemiol., 2021, 6:e07 [Crossref], [Google Scholar], [Publisher]
- Farnood F., Ren. Endocrinol., 2020, 7:e05 [Google Scholar], [Publisher]


