Document Type : Original Article
Authors
- Andrew Adhytia Lieputra 1, 2, 3
- Rebriarina Hapsari 1, 4, 5
- Cecilia Oktaria Permatadewi 1, 2
- Irfan Kesumayadi 1
- Iva Puspitasari 1, 2
- Purnomo Hadi 1, 2
- Nur Farhanah 1, 2
- Hendro Wahyono 1, 2
- Hery Djagat Purnomo 1, 2
1 Faculty of Medicine, Universitas Diponegoro, Semarang, Indonesia
2 Kariadi Hospital, Semarang, Indonesia
3 School of Medicine and Health Sciences, Atma Jaya Catholic University of Indonesia, Jakarta, Indonesia
4 KRMT Wongsonegoro Hospital, Semarang, Indonesia
5 Diponegoro National Hospital, Semarang, Indonesia
Abstract
Transmission prevention is important to prevent the spread of COVID-19. Although most cases are transmitted through droplets and aerosols, several studies have shown the possibility of transmission through fecal material. It is important to identify which patients are more likely to shed SARS-CoV-2 to raise awareness of the virus transmission via their feces. This study aims to determine the association of clinical and laboratory characteristics of COVID-19 patients with the SARS-CoV-2 detection in feces. From May to December 2020, fecal specimens from confirmed COVID-19 patients were collected, processed, and tested for the SARS-CoV-2 RNA presence. Clinical and laboratory parameters were compared between patients with and without SARS-CoV-2 RNA in their feces. Categorical variables were analyzed using Chi-square or Fisher’s Exact test, whereas non-categorical variables were analyzed using Independent T test and Mann-Whitney U test. From 51 COVID-19 patients of whom fecal specimen were collected, SARS-CoV-2 RNA was found in the feces of 26 (50.9%). The SARS-CoV-2 presence in the feces was associated with cough (p=0.002), dyspnea (p=0.017), bilateral pneumonia (p=0.011), lower SARS-CoV-2 CT-values in nasopharyngeal and oropharyngeal swabs (p=0.015), and clinical severity (p=0.0023). In conclusion, several clinical characteristics contributing to the COVID-19 severity and higher SARS-CoV-2 viral load in the respiratory tract were associated with the SARS-CoV-2 RNA detection in feces.
Graphical Abstract
Keywords
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the virus causing COVID-19 pandemic since early 2020 [1]. An evaluation of disease transmission is required to prevent disease spreading. Predominantly, COVID-19 is transmitted via respiratory tract secretions, but the SARS-CoV-2 RNA presence in other specimens such as blood, urine, genital secretion, and feces raised a possibility of other means of disease transmission [2–4].
Previous study showed that genetic material and viable SARS-CoV-2 have been isolated from fecal material [5, 6]. SARS-CoV-2 has been suspected with the possibility of transmission through fecal aerosols [7, 8]. In the previous SARS-CoV epidemic in Hong Kong in 2003, a case of super-spreading event in Amoy Garden Housing-Complex was hypothesized that virus was aerosolized through the sanitary plumbing [9]. Moreover, previous evidence showed that Corona Virus can survive in the environment in the sewage for few hours and up to 14 days [10]. This event might have a chance for recurring in SARS-CoV-2 epidemic. Furthermore, Angiotensin converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2), the receptor and co-receptor for SARS-CoV-2, are found in significant amounts in the intestine, increasing the potential for fecal transmission [11].
Environmental factor plays a pivotal role in disease transmission. Fecal transmission of SARS-CoV-2 would pose a huge challenge in developing and underdeveloped countries. Previous studies in Brazil and India have detected SARS-CoV-2 RNA in wastewater which correlated with the number of COVID-19 patients in the surrounding area [12, 13]. Meanwhile, Indonesia, where open defecation remains habitual in some areas, still has problems with population density living in urban areas, low hand washing skills, and low access to clean water [14]. In addition, hospitalized patients with severe COVID-19 should take special care while disposing of their waste, including feces, to prevent the virus from becoming aerosolized in the plumbing system. Therefore, the identification of clinical and laboratory characteristics that associated with the SARS-CoV-2 presence in feces is essential to controlled disease transmission, particularly via fecal materials.
Marteials and Methods
From May to December 2020, we collected fecal specimens from laboratory confirmed COVID-19 patients, aged >18 years old, who were admitted to Dr. Kariadi Hospital, Semarang, Indonesia. Patients were not recruited if they did not defecate during their admission, or the authority (patient or guardian) did not give consent to participate in this study.
The fecal material was collected at the earliest defecation after COVID-19 was confirmed by qRT-PCR test. Feces were processed by diluting it in Tris-HCl buffer, vortexed, and centrifugated. Supernatant was collected and stored at -80 °C until further use.
At the end of the study period, stored supernatant was extracted using DaAn Gene RNA/DNA Purification Kit Magnetic Bead (DaAn Gene Co., Ltd. of Sun Yat-Sen University, Guangzhou, China). The SARS-CoV-2 qRT-PCR was performed using DaAn 2019-nCoV Diagnostic kit regular (DaAn Gene Co. Ltd., Sun Yat-Sen University, Guangzhou, China) that targeted ORF1ab and N genes of SARS-CoV-2 and Human RNase P as internal control. The qRT-PCR interpretation was based on the manufacturer’s instructions.
Clinical characteristics such as symptoms and signs, comorbidities, chest X-ray, severity, and outcomes were examined in this study. The illness severity was determined based on criteria published by the Ministry of Health Republic Indonesia, which consist of asymptomatic, mild, moderate, severe, and critical [15]. Data on outcome variables, i.e. length of stay and mortality were further collected. Laboratory characteristics, i.e. complete blood count, blood chemistry (AST, ALT, albumin, urea, creatinine, and total bilirubin), inflammatory markers (CRP and procalcitonin level), CT values of naso/oropharyngeal, and fecal specimens were also examined.
Statistical analysis
Categorical variables were analyzed using Chi-square, or Fisher’s Exact test (when Chi-Square condition is not fulfilled). Independent T test was used to compare means between two groups if the data suggest for normal distribution, whereas Mann-Whitney U test was used when the data is not normally distributed. Spearman correlation was used to analyze between categorical and ordinal variables. All statistical analyses were performed with SPSS software (version 25.0, SPSS, Inc.). P-value <0.05 was considered significant.
Ethics statement
This study has been approved by the Institutional Review Board of the Dr. Kariadi Hospital, Semarang, Indonesia (Number 539/EC/KEPK-RSDK/2020)
Results and Discussion
This study enrolled 54 subjects who had laboratory confirmed COVID-19 and hospitalized during May to December 2020. Non-detection of internal control in 3 fecal samples, despite repeated SARS-CoV-2 qRT-PCR in these respective samples, left 51 subjects for analysis. Among these 51 subjects, 26 (50.9%) showed fecal presence of SARS-CoV-2 RNA with CT value of 34.58 (31.35-36.4).
Clinical characteristics of subjects
The median age of the subjects was 52 years old, ranging from 26-79 years old. Twenty-nine (56.9%) subjects were female. Cough (58.8%), fever (56.9%), and dyspnea (49.0%) were the most reported symptoms. Cardiovascular (33.3%) and diabetes (31.4%) were the top comorbidities. Cough and dyspnea were significantly associated with the SARS-CoV-2 RNA presence in the feces, P=0.002 and P=0.017, respectively. From the chest X-Ray, 7 subjects (13.7%) showed no abnormalities, whereas 2 (3.9%) and 42 (82.4%) revealed unilateral and bilateral pneumonia, respectively. All but one subject with detected SARS-CoV-2 RNA in feces had bilateral pneumonia (P=0.011). In addition, the increased COVID-19 severity was correlated with the SARS-CoV-2 RNA detection in feces (P=0.003). The Clinical characteristics of subjects are listed in Table 1.

Some previous studies showed no association between coughing with the SARS-CoV-2 RNA detection in feces [16-18]. In our study, we found an association of coughing with the SARS-CoV-2 RNA detection in feces. COVID-19 patients who have a cough might be unable to expectorate the respiratory secretion, which could eventually result in the virus being ingested. Pulmonary tuberculosis is an example of a respiratory disease that also is presented with cough symptoms, in which the nucleic acid of its etiologic pathogen can be detected in the feces and gastric lavage [19].
Bilateral pneumonia suggests more lung volume affected by SARS-CoV-2 infection. The bigger affected lung volume might result in a bigger pulmonary secretion resulting from lung cell destruction, leukocyte migration, and mucus production. Previous studies showed no association of bilateral affected lungs with the presence of SARS-CoV-2 in feces [17, 20].
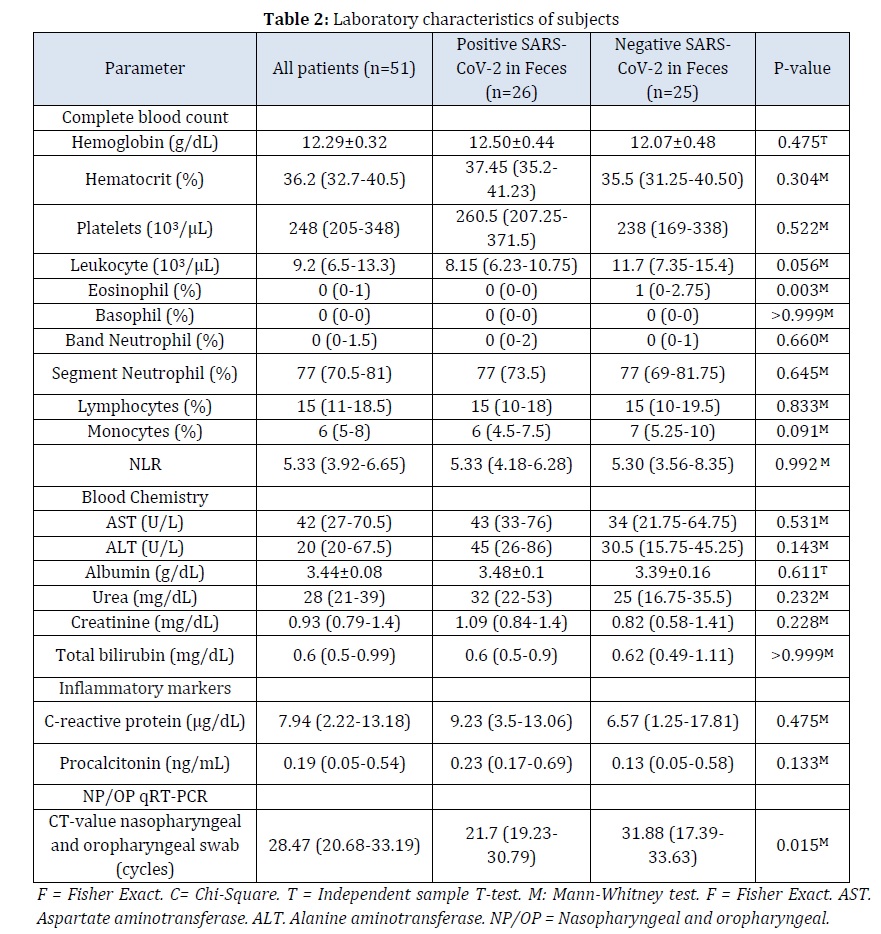
Laboratory characteristics
The laboratory characteristics of subjects is shown in Table 2. There was no difference in complete blood count parameters between two groups, except for eosinophil count (P=0.003). However, the eosinophil counts in both patient subsets, with or without RNA detection in feces, were within normal limits. There was also no difference in blood chemistry and inflammatory markers. NLR and CRP levels, markers for ongoing inflammation, in both patient subsets were increased. These are common findings for hospitalized COVID-19 patients [21, 22]. Positive SARS-CoV-2 RNA in feces has CT value of 34.58 (31.35-36.4). This finding is similar to a previous study that showed an average CT value of 33.4 ± 3.4 calculated from 88 samples [23]. In this study, positive detection of RNA in feces was associated with lower CT value of nasopharyngeal and oropharyngeal (NP/OP) swab (P=0.015). However, a previous study found no association between viral load in feces and NP/OP with R2 = 0.01, F (1, 52) = 1.92, p = 0.17 [24].
A lower CT value indicates a larger viral load, which increases the likelihood of ingesting the virus. Subsequently, enterocytes infection is possible because these cells also express the ACE-2 receptor [25-27]. Although the enterocyte can be infected, it is shown that the inflammatory evidence is low despite the presence of humoral response [28]. This finding supports that gastrointestinal symptoms may be absent in individuals with detected SARS-CoV-2 in feces.
It is still debatable whether SARS-CoV-2 in feces poses a high risk of COVID-19 transmission.
Several studies found viable viruses in the feces [4-6]. One study documented possible fecal aerosol transmission from index patients to other individuals sharing the drainage system that might be due to the U-shaped water trap dry out [8]. In contrast, other studies suggest that the fecal-oral route is highly unlikely because they could not retrieve viable or infectious SARS-CoV-2 despite low CT value of SARS-CoV-2 RNA in feces [29] and the use of concentration method [30]. A study in animal models demonstrated SARS-CoV-2 infections in ferrets after being inoculated with a fecal sample from a COVID-19 patient despite a failure in isolating viable virus in Vero cells from the original fecal sample [31].
Subjects’ outcome
The median of length of stay among participants in this study was 13 (10-20) days. Six (11.8%) subjects died, of whom 5 (19.2%) showed positive SARS-CoV-2 in feces. There was no difference in length of stay and mortality.
The present study has several limitations. First, the present study did not explore SARS-CoV-2 in sputum, gastric lavage, and gastrointestinal biopsy so that the SARS-CoV-2 association in ingested sputum that reached intestine could not be explored. Second, we did not perform SARS-CoV-2 culture from our fecal samples. Thus, we could not determine the SARS-CoV-2 viability in feces among our patient’s subsets. However, the present study has advantages, i.e. this study included more diverse clinical presentations ranging from asymptomatic to severe. This study further showed that SARS-CoV-2 RNA in fecal specimen stored in -80 °C did not affect the quality of extracted RNA.
Conclusion
In summary, our study demonstrated that several clinical characteristics (cough, dyspnea, and bilateral pneumonia) and laboratory (lower CT value of NP/OP specimen) contributing to the COVID-19 severity and higher SARS-CoV-2 viral load in the respiratory tract were associated with the SARS-CoV-2 RNA detection in feces. Exploration of the presence and viability of infectious SARS-CoV-2 in various levels of gastrointestinal tract and wastewater is required.
Acknowledgements
The authors would like to thank the Institute for Research and Community Services of Diponegoro University who grant this research (No. 233-60/UN7.6.1/PP/2020). We would also thank to the PT. Bakti Energi Abadi for the donation of RNA extraction kit and qRT-PCR kit. Finally, the authors would like to thank the research subjects who volunteered to participate and the doctors and nurses taken care the subjects at the inpatient isolation ward.
Disclosure Statement
No potential conflict of interest was reported by the authors.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors’ contributions
AAL and RH designed the study, submitted a research proposal, performed laboratory work, and wrote the manuscript. COP and NF examined the research subjects and wrote the manuscript. IK contributed on data analysis and wrote the manuscript. IP, HW, and HDP contributed to the manuscript writing. The final manuscript was reviewed and approved by all authors.
Orcid
Andrew Adhytia Lieputra
https://orcid.org/0000-0002-8425-6470
Rebriarina Hapsari
https://orcid.org/0000-0001-8367-063X
Cecilia Oktaria Permatadewi
https://orcid.org/0000-0001-8462-3018
Irfan Kesumayadi
https://orcid.org/0000-0001-7517-0942
Iva Puspitasari
https://orcid.org/0000-0003-3591-9255
Purnomo Hadi
https://orcid.org/0000-0003-4047-9418
Nur Farhanah
https://orcid.org/0000-0002-7385-3325
Hendro Wahyono
https://orcid.org/0000-0001-6234-8772
Hery Djagat Purnomo
https://orcid.org/0000-0003-4235-8564
HOW TO CITE THIS ARTICLE
Andrew Adhytia Lieputra, Rebriarina Hapsari, Cecilia Oktaria Permatadewi, Irfan Kesumayadi, Iva Puspitasari, Purnomo Hadi, Nur Farhanah, Hendro Wahyono, Hery Djagat Purnomo. Clinical and Laboratory Characteristics of COVID-19 Patients with Fecal SARS-CoV-2 Detection at a Tertiary Hospital in Indonesia. J. Med. Chem. Sci., 2023, 6(9) 1935-1942


