Document Type : Original Article
Authors
Biology Department, College of Science, University of Mustansiriyah, Baghdad, Iraq
Abstract
The enzymes extended spectrum β-lactamases (ESBLs) can hydrolyze oxyimino-cephalosporins. The abuse of new antibiotics leads to the emergence of new types of β-lactamases enzymes; because of the emergence of these reasons, this research aims to investigate Klebsiella pneumoniae-producing ESBLs in patients with urinary tract infection and determine the antibiogram pattern furthermore detect the frequency of mutations in the ctx-m gene. Fifty bacterial isolates of K. pneumoniae were isolated and identified phenotypically and chemically from patients with UTI. All isolates were subjected to an antibiogram using the disc diffusion method. MIC for cefotaxime antibiotic was carried out by microdilution method to all resistant isolates to cefotaxime disc; furthermore, screening of ctx-m gene was carried out by conventional PCR. Sequencing of the ctx-m gene and its mutations was done by the dideoxynucleotide sanger method and BioEdit software, respectively. Fifty bacterial isolates confirm K. pneumoniae; these isolates appeared antibiogram diversity to seventeen antibiotics ranging from high resistance to Ampicillin, Cefazolin, Ceftriaxone, Ampicillin/Sulbactam, Cefepime, and doxycycline while moderate resistance to ciprofloxacin, Moxifloxacin, and Cefotaxime. Furthermore, there is low resistance toward Tobramycin, gentamicin, Amikacin, Meropenem, Imipenem and no resistance toward Ertapenem. The MIC of most resistant isolates is more than 256μg/ml of cefotaxime. The PCR detection revealed that twelve of twenty-three isolates harbored the ctx-m gene, while the results of sequencing showed that many mutations occurred. Belong to a high incidence of infection with bacteria K. pneumoniae producing ESBL in the studied population, molecular studies in the field of antimicrobial resistance of bacteria become emerging to eradicate and prevent the spread of resistance strains.
Graphical Abstract
Keywords
Introduction
Due to the fast rise and expansion of antibiotic resistance by bacterial isolates, it is critical to track antibiotic usage and establish treatment options to reduce antibiotic misuse. As a result, it is critical to keep researching genes involved in antibiotic resistance in pathogenic bacteria, especially genes involved in extended-spectrum B-lactamase enzymes [1]. Klebsiella pneumoniae is a common source of infections in hospitals and the population. β-lactam antibiotics are widely utilized and efficient in treating MDR bacteria. The clinical isolates of bacteria K. pneumoniae are resistant various antibiotics, including B-lactam antibiotics that have been reported on several occasions [1, 2]. Although K. pneumoniae is a normal part of the intestinal flora, it can play a role in disorders including urinary tract infection, purulent infections, and septicemia.
The ability of k. pneumoniae to acquire resistant genes is significantly enhanced [3, 4]. It is an opportunistic microbe that can cause nosocomial infections, particularly in patients in critical care units and immune compromised. Furthermore, these bacteria can acquire antibiotic-resistant genes, which account for the pathogen's enhanced pathogenic capacity and the emergence of severe disease types [5]. The most prevalent mechanism to of Beta-lactam antibiotics was the production of Beta-lactamase enzymes. The problem of antibiotic resistance has been risen in medical practices because of the extended-spectrum beta-lactamases (ESBLs) [6, 7]. These enzymes carried by plasmid can hydrolyze lactam ring bonds from antibiotics such as pencillinis, cephalosporins, and aztreonam groups, and they are inhibited by inhibitors like clavulanic acid [8]. There is a risk of resistance to additional antibiotic families, like aminoglycosides, fluoroquinolones, trimethoprim-sulfamethoxazole, and tetracycline, because of the presence of many various modifying enzymes on the same plasmid [9]. The World Health Organization (WHO) included K. pneumoniae, which produces extended-spectrum beta-lactamase (ESBLs), in its list of the most hazardous superbugs in 2017 [10].
The extended-spectrum antibiotics like cephalosporin and monobactam can be hydrolyzed by extended-spectrum beta-lactamases enzymes. Cefotaximases (CTX-M), Temoneria (TEM), and Sulphydryl variable (SHV) are the main categories of these enzymes [11]. CTX-M genotypes have a significant activity toward ceftriaxone and cefotaxime than ceftazidime, and they have become the most extensively dispersed and internationally dominant genotypes. It consists of about 170 allelic variations that are segregated into five primary categories depending on amino acid sequence similarity: CTX-M1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25. Each category has many distinct variations, with dominant variants spread in particular geographic areas [12].
The importance of this research is to highlight the prevalence rate of the ctx-m gene among local ESBLs K. pneumoniae and to determine the types of mutations.
Material and Methods
Isolation and identification of K. pneumoniae
Fifty (50) bacterial isolates of K. pneumoniae were recovered from patients suffering from urinary tract infections who attended Baghdad hospitals from September 2021 to January 2022. The isolates were cultivated on blood and MacConkey media (HiMedia, India). Bacterial identification was made using classical methods such as morphological; biochemical tests and confirmed identification with Vitek 2 system (BioMeriuex, France) with ID-GNB cards depending on manufacturing instructions.
Antibiotics sensitivity test
Antimicrobial sensitivity towards seventeen antimicrobial agents was screened by subjecting all fifty isolates of K. pneumoniae to disc diffusion method with utilized (MHA) (HiMedia, India) depending on the CLSI guidelines [13]. Ceftriaxone (CRO 30 μg), Cefotaxime (CTX 30μg), Cefepime (CPM 30 μg), Cefazollin (CZ 30μg), Ampicillin (AMP 10 μg), Ampicillin/Sulbactam (SAM 10/10 μg), Etrapenem(ETP 10 μg),Imipenem (IMP 10 μg), Meropenem (MEM 10 μg), Amikacin (AK 30 μg), Gentamicin (GM 10 μg), Tobramycin (TOB 10 μg), Ciprofloxacin (CIP 5 μg), Doxcycline (DXT 30 μg),Moxifoxacin (MXF μg), Nitrofurantion (300 μg) and trimethoprim/sulfamethoxazole (SXT 1.25/23.75 μg). According to antibiogram activity, the dendrogram was built by using UPGAM pairwise.
Cefotaxime minimum inhibitory concentration
MIC for Cefotaxime was carried out using standard microdilution minimal inhibitory concentration method for isolates resistant to Cefotaxime disc and depending on the CLSI criteria [13]. Belonging to the CLSI guidelines, the breaking point of MIC values was (≥ 4 μg/mL). The experiment was done with two replicates and a standard strain of E. coli ATCC 25922 (CPHL, Baghdad). MIC was represented as the minimal concentration of antibiotics that inhibit bacterial division; as a result, absence of bacterial growth in media.
Molecular detection of ctx-m gene
The MDR K.pneumoniae were screened for the ctx-m gene by PCR technique. Firstly, genomic DNA was extracted according to the manufacturer’s instructions by (Wizbio/ Korea). The DNA purity and concentration were measured by Nanodrop (Thermo Scientific Nanodrop Lite UV visible Spectrophotometer/ USA). The PCR mixture was prepared by adding 12.5 μl of 2X GO taq green master mix (promiga/USA), 1.5 μl (0.6 pmol) from each universal primer with oligonucleotides sequence Forward 5`-SCSATGTGCAGYACCAGTAA-3` and Reverse CCGCRATATGRTTGGTGGTG-3`, 3 μl from template DNA and the addition of deionized nucleus free water was to complete the volume up to 25 μl. At the same time, the PCR process was done by using a thermocycler instrument (Tech-NET-500/USA) within amplification conditions contained: initial denaturation at (95°C/5 min.) followed by 35 cycles of initial denaturation at (95 °C/ 30 sec.), annealing at 58°C/30 sec.), extension at (72 °C/30 sec.) and final extension at (72 °C/10 min). The products of PCR amplification were visualized by subjecting the products to an electrophoresis process (Cleaver scientific/ Taiwan) using an agarose gel concentration of 1% and EtBr stain. And then envision under a UV transilluminator (U.V.P./USA). The twelve PCR products were sent to Macrogen company/ in Korea for detecting nucleotides sequence using sanger’s sequencing; furthermore, the data has been submitted to the NCBI database to analyze the mutations.
Results and Discussion
Isolation and identification
Fifty clinical isolates of K. pneumoniae were recovered from many hospitals in Baghdad city. All recovered isolates were from patients suffering from urinary tract infection; furthermore, there were identified phenotypically, chemically, and VITEK system as K. pneumoniae.
Antimicrobial sensitivity test
All isolates showed diversity in antibiogram resistance activity. The highest resistance was shown to be toward beside beta-lactam groups Ampicillin (49/50 %98), (45/50 %90) toward Cefazollin while forty-six isolates (%88) for each Ceftriaxone and Ampicillin/Sulbactam, forty isolates resistant (%80) toward each of Cefepime and Trimethopime/ sulfamothaxazole. Furthermore, thirty isolates resistant (60%) to quinolones antibiotics Ciprofloxacin and moxifloxacin (23/50 46%) toward Cefotaxime, isolates appeared resistant toward aminoglycoside group (15/50 30%, 14/50 28%, 3/50 6%) Tobramycin, Gentamicin and Amikacin respectively, (8/50 %16) toward Nitrofuranation, the carbapemen group (2/50 96%, 1/50 98%) isolates were resistant to Meropenem and Imipenem respectively the most effective antibiotic was Etrapenem and Doxycycline (0/ 50 100%) as shown in Figure 1.

Figure 1: Antibiogram sensitivity of K. pneumoniae
The isolates revealed high diversity referred to antibiogram activity, as shown in Figure 2.
Cefotaxime MIC
Twenty-three K. pneumoniae isolates that exhibited resistance to cefotaximeantibiotic disc diffusion method was evaluated by microdilution minimal inhibitory concentration test. All isolates showed high levels of resistance towards cefotaxime antibiotic, there was noticeably exhibited MIC > 64 μg/mL.
Molecular detection of ctx-m gene and sequencing
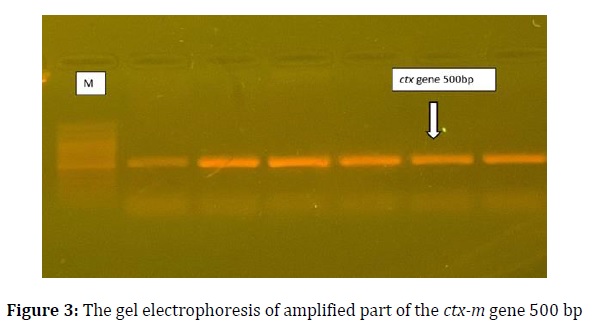
PCR amplification results showed that twelve isolates out of twenty-three (52.1%) carried the ctx-m gene; the ctx-m gene with 500 bp size appeared (Figure 3). The sequencing of PCR products to the ctx-m gene was submitted under accession number (LC712924-LC712935). The sequencing appeared many mutations occurred in the ctx-m gene in local isolates compared with the NCBI database. Bioedit software showed the mutations after aligning with a reference sequence in the Gene Bank, as shown in Table 1.
UTI infections are usually caused by Enterobacteriaceae, the first-line treatment frequently utilized by beta-lactam antibiotics [1]. The ESBLs enzyme production by Enterobacteriaceae varies widely over the world. The high frequency of MDR isolates in Enterobacteriaceae in all nations where patients are treated with antibiotics. North America and Europe had low rates, whereas Asia, South America, and Africa had high rates.


Antimicrobials obtained in the absence of a prescription, low hygiene, copy pharmaceuticals, a high frequency of infectious illnesses, and a deficiency in diagnostic tools for infections and antibiotic resistance all contribute to their spread [14]. K. pneumoniae-producing ESBLs have been investigated in the current study in UTI patients who attended Baghdad hospitals. The current study showed a high resistant rate toward many antibiotics and emerging MDR isolates, as shown in Figure 1; these findings were compatible with [1, 2]; they mentioned the high resistance rate in K. pneumoniae in Baghdad; these rates vary between many studies in Baghdad regarding the infection of through which bacteria isolated and laboratory techniques, moreover, we notice similar antibiotics resistance patron for many antibiotics used in a study created in Erbil/ Iraq [15]. The isolates showed high diversity belonging to dendrogram; many isolates were segregated to gather. The MIC of cefotaxime antibiotics showed a high-rate of resistance; most isolates appeared ≥16 μg/mL. The rate of resistance to third-generation cephalosporin antibiotics varies depending on countries and geographical regions [16]. PCR detection appeared in 52.1% of isolates harbored with the ctx-m gene in local isolates, while Erbil reached %41.1 [15]. Many studies conducted higher rates of prevalence of the ctx-m gene [12, 14, 16]. Many mutations have appeared in the ctx gene; types of these mutations diverse belong to geographical regions [16].
Conclusion
We found that the gene ctx-m is widely distributed in Iraqi isolates and these strains resist various antibiotics. The distribution rate of the ctx-m gene discovered in our investigation differed from that found in other countries, indicating significant diversity in the epidemiology of these resistance determinants and emphasizing the necessity of tools to monitor them in diverse epidemiological cases. To further comprehend the significance of vectors contra clonal spread in the distribution of these resistance determinants, researchers should look at the role of genetics in support and transferability of the different ESBL determinants and the clonal diversity of isolates.
Acknowledgments
The authors would like to express their gratitude, and they are thankful to Mustansiriyah University (www.Mustansiriyah.com) for supporting the process and completion of this study.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors' contributions
All authors contributed to data analysis, drafting, and revising of the paper and agreed to be responsible for all the aspects of this work.
Conflict of Interest
The author declared that they have no conflict of interest.
ORCID:
Hadeel Musafer
https://orcid.org/0000-0003-4783-7177
Majid Al-Bayati
https://orcid.org/0000-0002-1366-6405
Sawsan Kareem
https://orcid.org/0000-0003-2627-8716
HOW TO CITE THIS ARTICLE
Hadeel Musafer, Majid Al-Bayati, Sawsan Kareem. Epidemiology and Genetic Diversity of CTX-M Gene in Klebsiella Pneumoniaein Baghdad City. J. Med. Chem. Sci., 2023, 6(6) 1419-1425


