Document Type : Original Article
Authors
1 Department of Chemistry, College of Education for pure Science, University of Kerbala, Iraq
2 Department of Chemistry, Faculty of Science, University of Kufa, Iraq
Abstract
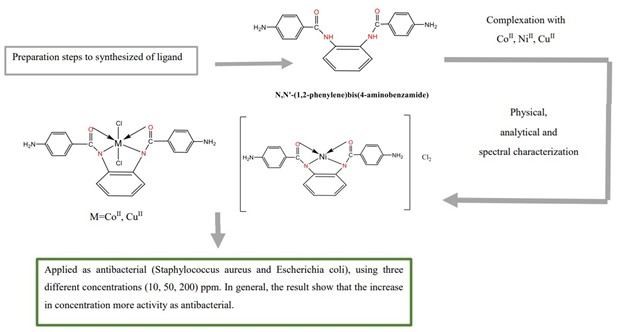
By condensation reaction between P-amino benzoic acid and ethanol in the presence of concentrated H2SO4 to produce ethyl 4-aminobenzoate, which was then converted into N, N'-(1,2-phenylene) bis(4-aminobenzamide) type [H2L] (N2O2) product by direct interaction between the two reactive molecules of this product with one molecule of 1,2-Benzene diamine, as a new series of metal conversion complexes of Co(II), Ni(II), and Cu(II). The prepared ligand and their complexes characterized in the solid and solution states using spectroscopic techniques such as UV-Vis, FT.IR, 1HNMR, and the elemental analyses as well as melting temperatures, molar conductivity, and magnetic susceptibility. The octahedral geometry around the central metal is suggested by molar conductivity, magnetic moment, molar ratio, and spectroscopic data about copper and cobalt ions, while tetrahedral around the nickel ion. After the incubation period at 37 °C for 24 h, the biological behavior of the binding produced with its anti-bacterial compounds against (Staphylococcus aureus and Escherichia coli) was examined at different concentrations (10, 50, and 200) ppm. The results showed that the performance of the prepared compounds was better in resisting and reducing the growth of bacteria tested at high concentrations.
Graphical Abstract
Keywords
Main Subjects
Introduction
Amides are a type of functional group which can be found in both synthetic and natural chemical compounds [1]. It is essential to take life seriously [2]. Amide grouping is a fundamental activity carried out by organic matter seen in common natural compounds such as proteins. It is stable and polar [3]. Amide bonds are commonly utilized in organic synthesis, medicinal chemistry, and agrochemicals, but they can also be used in other fields [4-6]. It is molecule acts as potent metal ion chelate, due to its oxygen and nitrogen atoms [7,8].
This notch can also be found in many synthetic substances, such as active pharmaceutical components or pro-drugs [9-13]. A diverse range of proteins, chemicals, medicines, and bimolecular structures use the amide process [14]. The amide group is a popular issue in drug research because of these factors [15].
The classic procedure for the synthesis of amides is the reaction of carboxylic acids with amines, usually in the presence of coupling reagents, or with carboxylic acid derivatives, esters, anhydrides, acyl halides, and the other activated species in the presence of the third amine [16]. Formalization of ester or amide bonds occur between an acid and a condensed alcohol or amine, respectively [17].
Amides are used in a variety of applications, including the employment of potentially hazardous chemicals or catalysts, medicinal development, and the polymer sector [18-19]. Despite the lack of evidence for amide synthesis from cinnamic derivatives, amides derived from cinnamic acids have been shown to exhibit antioxidant, anti-atherosclerotic, anti-fungal, cytotoxic and anti-viral properties [20,21]. There are other modern methods for preparing amide compounds [22].
Materials and Methods
Chemicals were supplied by Merck, Sigma-Aldrich and Fluka and used as received. 4-aminobenzoic acid, 1,2-benzene diamine, absolute ethanol, sulfuric acid, hexane, ethyl acetate, DMSO, and methanol, CuCl2.2H2O, NiCl2.6H2O, CoCl2.6H2O.
Apparatus
The melting point of compounds produced was measured by using Gallen kamp melting point, in the University of Karbala. The elemental microanalysis was performed using the Euro EA3000, in the University of Babylon. The molar conductivity for complexes was measured using inluba WTW balance, in the University of Karbala. The magnetic susceptibility of prepared complexes was measured by using magnetic sufcetitility balance, in the University of AL-Nahrain.
Using tetra methyl silane as an internal standard and DMSO as a solvent, 1H-NMR spectra were collected on a JNM - model Joal 400 MHZ. At sapala organics private, measurements were taken (India). In the University of Karbala, FT.IR spectra were recorded on an FT.IR 8400s, schimadzu- spectrophotometer, and KBr and CsI discs were utilized to measure FT.IR spectra in the region of wavenumber 4000-200 cm-1. In a liquid state, Uv-Vis. 1800 PC was used to get the electronic spectra of the produced ligand with its metal complexes with wavelengths spanning from 1100 to 190 nm, Shimadzu spectrophotometer.
Synthesis of N, N'-(1,2-phenylene) bis(4-aminobenzamide) [L]
Prepare this ligand according to the following steps:
Preparation of ester ethyl 4-aminobenzoate
P-amino benzoic acid (0.152 g - 0.0018 mol) was dissolved in enough amount of the absolute ethanol (20 mL) and (1 mL) of concentration of H2SO4 was added, the mixture was refluxed for 3-5 h, and was cooled to (25 °C). The product was filtered and refined by recrystallization of the absolute ethanol.

Scheme 1: Synthesis of ester
Preparation of N, N'-(1,2-phenylene) bis(4-aminobenzamide) [L]
1,2-Benzene diamine (0.1 g, 0.001 mol) of added to ethyl 4-aminobenzoate (0.33 g ,0.002 mol) in (25 mL) of absolute ethanol, the blend of compounds was refluxed for 3 h, after cooling, the precipitate was filtered and washed with some amounts of cooled ethanol, then it was recrystallized from ethanol.

Scheme 2: Synthesis of ligand
Synthesis of L- metal complexes
Complexes were synthesized by the reaction of 1 mole of identical metal (CoCl2.6H2O, NiCl2.6H2O, and CuCl2.2H2O) which were dissolved in 5-10 mL of methanol, with a 1 mole of the ligand was dissolved in 30 mL of methanol. The reaction mixture was refluxed and stirring for 2 hours at 60 – 65 °C. The products were filtered, washed with ethanol, and desiccated. The physical produce data for the ligand and its complexes are listed in Table 1.

Scheme 3: Synthesis of complexes
The body's antibacterial activity
Antibacterial activity was investigated using agar diffusion [6]. The potency against (Escherichia coli and Staphylococcus aureus) was determined using a method which involved inoculating the surface of Moller-Hinton agar on a regular basis by spreading 100 L of (1×109 cells/ml) bacterial suspension into a Petri dish (90 mm diameter) for 24 hours, while adjusting the turbidity of the suspension. McFarland’s solution is used as a standard by bacteria. Immerse 0.1 ml of each concentration 10, 50 and 200 ppm of (the prepared ligand and its complexes [L-Co(II), L-Ni(II), and L-Cu(II)] dissolved in dimethyl sulfoxide) in Holes (5 mm in diameter) for half an hour on the plates. After a 24-hour incubation period at 37 °C, the anti-bacterial capacity of the plates was assessed by measuring the inhibitor.
Results and Dissections
The bonding was accomplished in two steps: first, P-aminobenzoic acid and ethanol was interacted with a high concentration of H2SO4 to generate an amino-benzoate complex, then 1,2-benzene diamine was added as an aminobenzamide and the complexes were formed by reacting metal ions with the synthesized ligand, as indicated in Scheme 1,2 and 3. The physical properties were summarized in Table 1. The analytical data of the microelements of the ligand and their complexes are listed in Table 2. These findings matched the predicted formula and were insoluble in non-polar or organic solvents, but soluble in dimethyl formamide (DMF), dimethyl sulfoxide (DMSO), methanol, and acetone. Except for nickel which is a non-electrolyte. Table 3 displays the molar electrical conductivity of the complexes in DMSO, suggesting that they are electrolytes. Table 4 summarizes the magnetic sensitivity as well as the ratio (1:1). The band mentioned at 3306.1 cm-1, which is related to the stretching υ (NH), and υ (C=O) stretching at 1645.33 cm-1, which is attrition, may be seen in the infrared spectra of the ligand [23].
The complexes appear to dissipate and the band (NH) vanishes due to coordination and frequency shifts (C=O) and the other ranges are summarized in Table 5. The (π π*) transition produces a broad peak at 260–300 nm, while the (n π*) transition produces a narrow peak at 420 nm in UV-Vis ligand spectra [24]. The charge transfers in the UV region, as well as the d-d transition for the Co+2 complex at 625 nm for the type 4T1g 4T1gP complex, are indicated, as the d-d transitions for the Ni+2 complex at 576 and 602 nm for the type 4T1 4T2 and 4T1 4T1P complex, and the d-d transition for the Cu+2 complex at 785 [25] show nm type 2E 2T2.
The recommended shape and electrical transitions the 1H-NMR spectra of ligands and their complexes, are listed in Table 6. In DMSO-d6, the chemical shift at δ = 9.351 ppm is ascribed to amide group protons, the aromatic ring protons occur at δ =7.212 - 7.589 ppm and the chemical shift at δ =5.483-5.254 ppm is assigned to amine group protons [26-27].
In the spectra of the complexes, the disappearance of the amide group protons is shown which indicates the coordination of the ligand with the cobalt ion via the donor atoms [28-30] of ligand all data was listed in Table 7 which depicted the complexes planned shapes.
Table 1: Chemical compounds’ physical characteristics
|
Compound |
m.p. (oC) |
M.Fo. |
M.Wt |
Color |
Yield % |
|
H2L |
298-300 |
C20H18N4O2 |
346.14 |
pale brown |
35 |
|
[CoLCl2] |
150dec. |
C20H16Cl2CoN4O2 |
474.21 |
Deep brown |
66 |
|
[Ni L]Cl2 |
230dec. |
C20H16Cl2NiN4O2 |
403.06 |
pale green |
45 |
|
[CuLCl2] |
117dec. |
C20H16Cl2CuN4O2 |
478.82 |
Deep blue |
50 |
Table 2: The elemental microanalysis
|
Compound |
C% |
H% |
N% |
O% |
||||
|
|
Cal. |
Exp. |
Cal. |
Exp. |
Cal. |
Exp. |
Cal. |
Exp. |
|
H2L |
69.38 |
69.35 |
5.26 |
5.24 |
16.19 |
16.15 |
9.27 |
9.25 |
|
[CoLCl2] |
50.69 |
50.66 |
3.42 |
3.39 |
11.83 |
11.80 |
6.77 |
6.74 |
|
[Ni L]Cl2 |
59.62 |
59.59 |
3.97 |
3.95 |
13.6 |
13.3 |
7.98 |
7.96 |
|
[CuLCl2] |
50.18 |
50.15 |
3.35 |
3.32 |
11.9 |
11.6 |
6.63 |
6.61 |
Table 3: The molar conductivity
|
Compound |
Λm (S.mol-1.cm2) In(DMSO) |
|
[CoLCl2] |
27.2 |
|
[Ni L]Cl2 |
73.6 |
|
[CuLCl2] |
21.8 |
Table 4: The magnetic susceptibility
|
Compound |
meff B.M |
|
[CoLCl2] |
3.65 |
|
[Ni L]Cl2 |
2.77 |
|
[CuLCl2] |
1.62 |
Table 5: The spectral data of the chemicals which have been synthesized
|
Compound |
υ (-NH2) |
υ (-NH) |
υ (C=O) |
υ(C-H) aromatic |
υ (M-O) |
υ (M-N) |
|
[H2L] |
3412.19 |
3306.10 |
1645.33 |
3037.99 |
- |
- |
|
[CoLCl2] |
3358.18 |
- |
1637.62 |
2972.40 |
628.81 |
511.15 |
|
[Ni L]Cl2 |
3331.18-3321.52 |
- |
1612.54 |
2910.68 |
638.46 |
578.68 |
|
[CuLCl2] |
3363.97 |
- |
1068.69 |
2989.76 |
623.03 |
538.16 |
Table 6 Electronic spectral data
|
Compound |
λ max nm |
λ max cm-1 |
Assigument |
Proposed structure |
|
[CoLCl2] |
629 |
15898 |
4T1g 4Tp1g |
O.h |
|
[Ni L]Cl2 |
576 602 |
17361 16611 |
4T1 4T2 4T1 4TP1 |
T.d |
|
[CuLCl2] |
785 |
12738 |
2E 2T2 |
O.h |
Table 7: 1H NMR spectral data
|
Compound |
(-NH2) |
() |
(C-H) aromatic |
|
[H2L] |
5.483-5.254 |
9.351 |
7.589-7. 212 |
|
[CoLCl2] |
5.357-5.302 |
- |
7.571-7.049 |

Scheme 4: The proposed geometry of synthesized complexes
Anti-bacterial activities
The ligand and its mineral complexes were evaluated in vitro against Staphylococcus aureus as Gram-positive and Escherichia coli as Gram-negative species at three different doses [31]. The data reveal that most of the complexes are more dangerous to these bacteria than the free ligand at higher concentrations.
According to the data, complexes have a significant influence on both gram-negative and gram-positive bacteria. The geometrical structure of these complexes, the nature of the metal ion, the nature of organic molecules, and the chelate effect, the nature of atoms which is coordinated with metals and the orientation of the ligand around the metal ion, the nature of metals and their oxidation states may all play a role.
Table 8: The diameters of the antibacterial activity of the ligand and the inhibitory area of its metal complex (mm)
|
Comp. |
Escherichia coli |
Staphylococcus aureus |
||||
|
E.coli |
Staphy. |
|||||
|
200 |
50 |
10 |
200 |
50 |
10 |
|
|
L |
20 |
17 |
15 |
18 |
16 |
12 |
|
L-Co(II) |
23 |
20 |
18 |
20 |
17 |
14 |
|
L-Ni(II) |
28 |
25 |
22 |
25 |
22 |
21 |
|
L-Cu(II) |
25 |
23 |
21 |
23 |
20 |
16 |
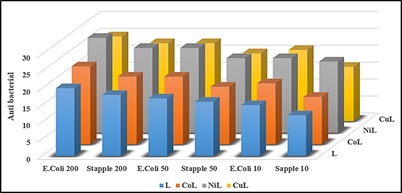
Figure 1: The compounds under research have antibacterial action
Conclusion
According to the findings, new N,N'-(1,2-phenylene) bis(4-aminobenzamide) and its complexes were synthesized and their structure was validated using spectroscopic testing at ambient temperature, elemental analysis, molar conductivity, and magnetic moment. In the solid state, we found that the Ni(II) complex has four coordination numbers with tetrahedral geometries, while the Co(II) and Cu(II) complexes have six coordination numbers with octahedral shape. These compounds were tested for antibacterial (Staphylococcus aureus, Escherichia coli) activities and the synergetic effect resulted in higher biological activity for these complexes than the free ligand.
Funding
This research did not receive any specific grant from fundig agencies in the public, commercial, or not-for-profit sectors.
Authors' contributions
All authors contributed toward data analysis, drafting and revising the paper and agreed to responsible for all the aspects of this work.
Conflict of Interest
We have no conflicts of interest to disclose.
ORCID:
Jihan Hameed Abdulameer
https://www.orcid.org/0000-0003-2850-8458
HOW TO CITE THIS ARTICLE
Jihan Hameed Abdulameer, Kadhim Hussein Serih, Hutham Abd Ali Abd Al Hussain Co(II), Ni(II) and Cu(II) Mononuclear Complexes with N2O2 Ligand Derived from Ethyl 4-Aminobenzoate, J. Med. Chem. Sci., 2022, 5(6) 874-880
- Boström J., Brown D.G., Young R.J., Keserü G.M., Nat Rev Drug Discov., 2018, 17:709 [Crossref], [Google scholar], [Publisher]
- Boyle M., Livingstone K., Henry M.C., Elwood J.M.L., Lopez-Fernandez J.D., Jamieson C., Lett., 2022, 24:334 [Crossref], [Google scholar], [Publisher]
- Gericke R., Wagler J., Chem., 2020, 59:6359 [Crossref], [Google scholar], [Publisher]
- Wachtler E., Gericke R., Block T., Pöttgen R., Wagler J., Chem., 2020, 59:15541 [Crossref], [Google scholar], [Publisher]
- Rodrigues R.R., Gabbaï F.P., Molecules, 2021, 26:1985 [Crossref], [Google scholar], [Publisher]
- Medeiros A.C.R.F., Gouveˆa M.M., Felipe T.V., Carvalho Marques F.F., Bernardino A.M.R., Ortiz F.L., Souza M.C., New J. Chem., 2019, 43:13881-13890 [Crossref], [Google scholar], [Publisher]
- Alizadeh R., Ghazinia N., J. Chem. A, 2019, 2:184 [Crossref], [Google scholar], [Publisher]
- Indira S., Vinoth G., Bharathi M., Bharathi K., Mol. Struct., 2019, 1198:126886 [Crossref], [Google scholar], [Publisher]
- Luo Y., Fan S., Yang J., J. Fang, Xu P., Dalton Trans, 2011, 40:3053 [Crossref], [Google scholar], [Publisher]
- Lu B., Xiao W.J., Chen J.R., Molecules, 2022, 27:517 [Crossref], [Google scholar], [Publisher]
- Czégéni C.E., De S., Udvardy A., Derzsi N.J., Papp G., Papp G., Joó F., Catalysts, 2020, 10:125 [Crossref], [Google scholar], [Publisher]
- Hua T.B., Yang Q.Q., Zou Y.Q., Molecules, 2019, 24:3191 [Crossref], [Google scholar], [Publisher]
- Wang P.Z., Zhao Q.Q., Xiao W.J., Chen J.R., Green Synthesis Catal., 2020, 1:42 [Crossref], [Google scholar], [Publisher]
- Jianga H., Studer A., Soc. Rev., 2020, 49:1790 [Crossref], [Google scholar], [Publisher]
- Bird R.E., Lemmel S.A., Yu X., Zhou Q.A., Bioconjugate Chem., 2021, 32:2457 [Crossref], [Google scholar], [Publisher]
- Sabatini M.T., Boulton L.T., Sneddon H.F., Sheppard T.D., Catal., 2019, 2:10 [Crossref], [Google scholar], [Publisher]
- Luu T.G., Jung Y., Kim H.K., Molecules, 2021, 26:7380 [Crossref], [Google scholar], [Publisher]
- Fan J., Wei Q., Zhu E., Gao J., Cheng X., Lu Y., Loh T.P., Commun., 2021, 57:5977 [Crossref], [Google scholar], [Publisher]
- Indira G., Vinoth M., Bharathi R., Chem. Intermed., 2021, 47:3051 [Crossref], [Google scholar], [Publisher]
- Jiaoyang Wang, Danfeng Wang and Xiaofeng Tong, Biomol. Chem., 2021, 19:5762 [Crossref], [Google scholar], [Publisher]
- Mohammadi A.A., Taheri S., Ghaderi P., Ahdenov R., Azizian H., Mohammadi-Khanaposhtani M, FaramarziA., Larijani B., Mahdavi M., J. Mol. Struct., 2021, 1227:129531 [Crossref], [Google scholar], [Publisher]
- Bering L., Craven E.J., Sowerby Thomas S.A., Shepherd S.A., Micklefield J., Commun., 2022, 13:1 [Crossref], [Google scholar], [Publisher]
- Mahmoudi F., Farhadi S., Dusek M., Poupon M., J. Chem. A, 2020, 3:534 [Crossref], [Google scholar], [Publisher]
- Alabdali A.J., IOSR JAC, 2013, 3:5 [Crossref], [Google scholar], [Publisher]
- Cortes-Clerget M., Akporji N., Zhou J., Gao F., Guo P., Parmentier M., Gallou F., Berthon J.Y., Lipshutz B.H., Commun., 2019, 10:2169 [Crossref], [Google scholar], [Publisher]
- Borja Cendon, Marc Font, Jose Mascareñas, and Moises Gulıas, ACS Catal., 2020, 10:3425 [Crossref], [Google scholar], [Publisher]
- Grishina G.V., Veselov I.S., Safronova E.N., Mazur D.M., Samoshin V.V., Tetrahedron Lett., 2017, 58:2019 [Crossref], [Google scholar], [Publisher]
- Noji M., Kondo H., Yazaki C., Yamaguchi H., Ohkura S., Takanami T., Tetrahedron Lette., 2019, 60:1518 [Crossref], [Google scholar], [Publisher]
- Koyama S., Kawai M., Takaishi S., Yamashita M., Hoshino N., Akutagawa T., Kanno M., Iguchi H., Crystals, 2020, 10:1081 [Crossref], [Google scholar], [Publisher]
- Angajalaa G., Arunaa V., Pavan P., Reddy P.G., Chem., 2022, 119:105533 [Crossref], [Google scholar], [Publisher]
- Vijayarohini P., Kavitha G., Alwar S.B.S., Swamidoss C.M.A., Today Proc., 2020, 33:4198 [Crossref], [Google scholar], [Publisher]